马闯等《Briefings in Bioinformatics》2021年
论文题目:Genome optimization via virtual simulation to accelerate maize hybrid breeding
论文作者:Qian Cheng, Shuqing Jiang, Feng Xu, Qian Wang, Yingjie Xiao, Ruyang Zhang, Jiuran Zhao, Jianbing Yan, Chuang Ma and Xiangfeng Wang
论文摘要:The employment of doubled-haploid (DH) technology in maize has vastly accelerated the efficiency of developing inbred lines. The selection of superior lines has to rely on genotypes with genomic selection (GS) model, rather than phenotypes due to the high expense of field phenotyping. In this work, we implemented ‘genome optimization via virtual simulation (GOVS)’ using the genotype and phenotype data of 1404 maize lines and their F1 progeny. GOVS simulates a virtual genome encompassing the most abundant ‘optimal genotypes’ or ‘advantageous alleles’ in a genetic pool. Such a virtually optimized genome, although can never be developed in reality, may help plot the optimal route to direct breeding decisions. GOVS assists in the selection of superior lines based on the genomic fragments that a line contributes to the simulated genome. The assumption is that the more fragments of optimal genotypes a line contributes to the assembly, the higher the likelihood of the line favored in the F1 phenotype, e.g. grain yield. Compared to traditional GS method, GOVS-assisted selection may avoid using an arbitrary threshold for the predicted F1 yield to assist selection. Additionally, the selected lines contributed complementary sets of advantageous alleles to the virtual genome. This feature facilitates plotting the optimal route for DH production, whereby the fewest lines and F1 combinations are needed to pyramid a maximum number of advantageous alleles in the new DH lines. In summary, incorporation of DH production, GS and genome optimization will ultimately improve genomically designed breeding in maize.
Short abstract: Doubled-haploid (DH) technology has been widely applied in maize breeding industry, as it greatly shortens the period of developing homozygous inbred lines via bypassing several rounds of self-crossing. The current challenge is how to efficiently screen the large volume of inbred lines based on genotypes. We present the toolbox of genome optimization via virtual simulation (GOVS), which complements the traditional genomic selection model. GOVS simulates a virtual genome encompassing the most abundant ‘optimal genotypes’ in a breeding population, and then assists in selection of superior lines based on the genomic fragments that a line contributes to the simulated genome. Availability of GOVS (https://govs-pack.github.io/) to the public may ultimately facilitate genomically designed breeding in maize.
传统育种主要依赖育种家的经验,基于候选材料的系谱关系以及田间的表型测定结果,进行选系和组配,存在随机性大,效率低下等问题。全基因组选择(Genomic selection,GS)被认为是一种极具潜力的育种新策略,可通过已知分子标记和表型的个体构建模型,估计出未测定表型个体的表型或育种价值以辅助早期选择,进而提升育种效率,缩短育种年限。然而,由于玉米杂交种中存在复杂的遗传互作与杂种优势效应,GS针对遗传力较低的数量性状(例如,产量)的预测效果并不令人满意。基于“优良基因越多,目标性状表现越好”的基本思想,该研究提出了“基因组优化设计”的策略,并开发了对应的R语言软件包GOVS,通过算法模拟虚拟基因组(Virtually optimized genome)尽可能多地聚合育种群体中目标表型的优势基因组片段或有利等位基因,并估算出每个育种材料对虚拟基因组的贡献情况,进而辅助育种家筛选合适的材料并进行后续的杂交实验。利用GOVS,进一步结合基因组选择以及双单倍体诱导(Doubled Haploid, DH)技术,可以指导育种家利用最少的遗传材料、最短的育种周期,最大限度地聚合优势等位基因,有望加速遗传增益的获取速度,加快玉米杂交育种进程。GOVS软件访问网址:https://govs-pack.github.io。
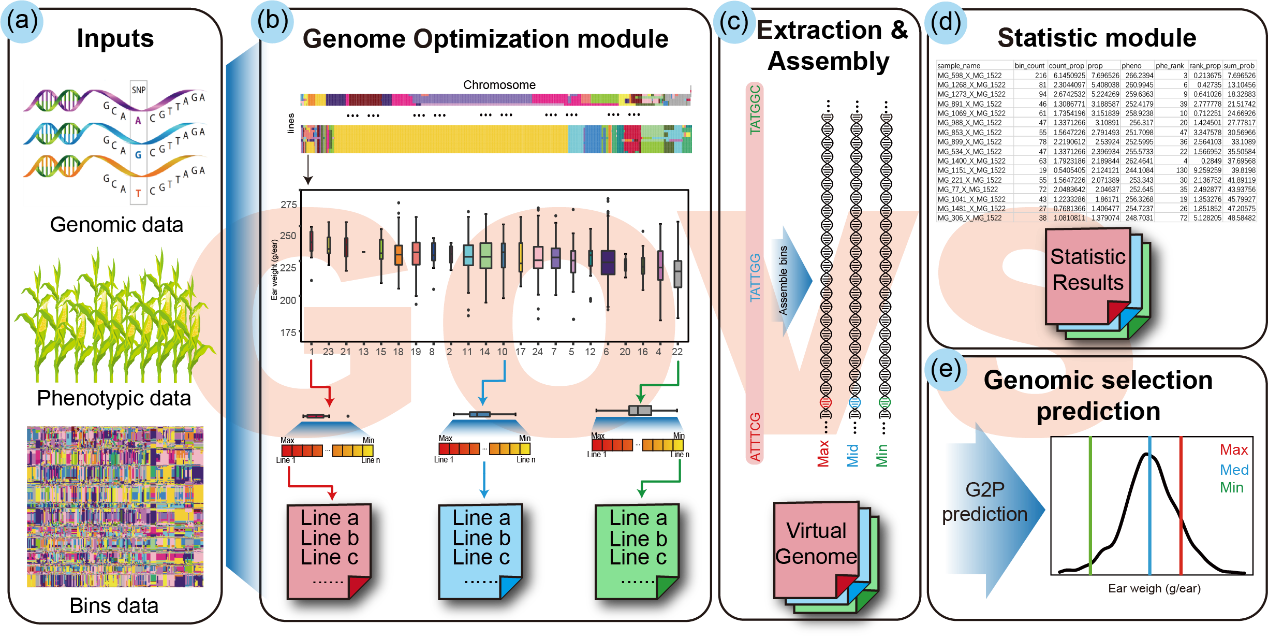
图1 GOVS 整体结构概览
伟德BETVLCTOR1946博士生程前以及中国农业大学农学院、国家玉米改良中心博士后姜淑琴为本文共同第一作者。伟德BETVLCTOR1946马闯教授以及中国农业大学农学院、国家玉米改良中心王向峰教授为论文的共同通讯作者。该论文受到国家自然科学基金等项目的资助。
文章链接:https://doi.org/10.1093/bib/bbab447
